Anthony Bretaudeau
Affiliations
Contributions
The following list includes only slides and tutorials where the individual or organisation has been added to the contributor list. This may not include the sum total of their contributions to the training materials (e.g. GTN css or design, tutorial datasets, workflow development, etc.) unless described by a news post.
Editorial Roles
This contributor has taken on additional responsibilities as an editor for the following topics. They are responsible for ensuring that the content is up to date, accurate, and follows GTN best practices.
- Topic: Genome Annotation
- Topic: GMOD
- Learning Pathway: Genome annotation for eukaryotes
- Learning Pathway: Genome annotation for prokaryotes
- Learning Pathway: Gallantries Grant - Intellectual Output 3 - Data stewardship, federation, standardisation, and collaboration
Tutorials
- Sequence analysis / Quality Control ✍️ 🧐
- Sequence analysis / SARS-CoV-2 Viral Sample Alignment and Variant Visualization 🧐
- Sequence analysis / Mapping 🧐
- Transcriptomics / De novo transcriptome assembly, annotation, and differential expression analysis ✍️
- Transcriptomics / De novo transcriptome reconstruction with RNA-Seq 🧐
- Transcriptomics / RNA-Seq analysis with AskOmics Interactive Tool ✍️ 🧐
- Transcriptomics / RNA Seq Counts to Viz in R 🧐
- Transcriptomics / RNA-seq Alignment with STAR 🧐
- Genome Annotation / CRISPR screen analysis 🧐
- Genome Annotation / Functional annotation of protein sequences ✍️
- Genome Annotation / Long non-coding RNAs (lncRNAs) annotation with FEELnc 📝 🧐
- Genome Annotation / Genome annotation with Funannotate ✍️ 🧐
- Genome Annotation / Genome annotation with Helixer ✍️ 🧐
- Genome Annotation / Bacterial genome quality control 🧐
- Genome Annotation / Bacterial pangenomics 🧐
- Genome Annotation / Genome annotation with Prokka 🧐
- Genome Annotation / Dataset construction for bacterial comparative genomics 🧐
- Genome Annotation / Bacterial Genome Annotation 🧐
- Genome Annotation / Genome annotation with Braker3 ✍️ 🧐
- Genome Annotation / Genome annotation with Maker ✍️ 🧐
- Genome Annotation / Comparative gene analysis in unannotated genomes 🧐
- Genome Annotation / Refining Genome Annotations with Apollo (eukaryotes) ✍️ 🧐
- Genome Annotation / Creating an Official Gene Set ✍️ 🧐
- Genome Annotation / Identification of AMR genes in an assembled bacterial genome 🧐
- Genome Annotation / Refining Genome Annotations with Apollo (prokaryotes) ✍️ 🧐
- Genome Annotation / Masking repeats with RepeatMasker ✍️ 🧐
- Genome Annotation / Genome annotation with Maker (short) ✍️ 🧐
- Foundations of Data Science / dplyr & tidyverse for data processing 🧐
- Foundations of Data Science / Introduction to SQL 🧐
- Foundations of Data Science / SQL with Python 🧐
- Foundations of Data Science / Make & Snakemake 🧐
- Foundations of Data Science / CLI basics 🧐
- Foundations of Data Science / SQL with R 🧐
- Foundations of Data Science / Plotting in Python 🧐
- Foundations of Data Science / Advanced R in Galaxy 🧐
- Foundations of Data Science / SQL Educational Game - Murder Mystery 🧐
- Foundations of Data Science / Advanced Python 🧐
- Foundations of Data Science / R basics in Galaxy 🧐
- Foundations of Data Science / Variant Calling Workflow 🧐
- Foundations of Data Science / Advanced SQL 🧐
- Foundations of Data Science / Advanced CLI in Galaxy 🧐
- Foundations of Data Science / CLI Educational Game - Bashcrawl 🧐
- Foundations of Data Science / Introduction to Python 🧐
- Teaching and Hosting Galaxy training / Running a workshop as instructor 🧐
- Teaching and Hosting Galaxy training / Asynchronous training 🧐
- Teaching and Hosting Galaxy training / Galaxy Admin Training 🧐
- Teaching and Hosting Galaxy training / Course Builder 🧐
- Using Galaxy and Managing your Data / InterMine integration with Galaxy 🧐
- Using Galaxy and Managing your Data / RStudio in Galaxy 🧐
- Development in Galaxy / Contributing to BioBlend as a developer 🧐
- Development in Galaxy / Creating Galaxy tools from Conda Through Deployment 🧐
- Development in Galaxy / Galaxy Interactive Tools ✍️ 🧐
- Assembly / Genome assembly using PacBio data ✍️ 🧐
- Assembly / Making sense of a newly assembled genome 🧐
- Assembly / Chloroplast genome assembly 🧐
- Assembly / Unicycler Assembly 🧐
- Assembly / Genome Assembly Quality Control ✍️ 🧐
- Assembly / Genome Assembly of a bacterial genome (MRSA) sequenced using Illumina MiSeq Data 🧐
- Assembly / Genome Assembly of MRSA from Oxford Nanopore MinION data (and optionally Illumina data) 🧐
- Introduction to Galaxy Analyses / Galaxy Basics for everyone 🧐
- Computational chemistry / High Throughput Molecular Dynamics and Analysis 🧐
- Visualisation / Genomic Data Visualisation with JBrowse 🧐
- Visualisation / Visualisation with Circos 🧐
- Ecology / Species distribution modeling 🧐
- Contributing to the Galaxy Training Material / Creating a new tutorial 🧐
- Contributing to the Galaxy Training Material / Including a new topic 🧐
- Contributing to the Galaxy Training Material / Creating content in Markdown 🧐
- Variant Analysis / M. tuberculosis Variant Analysis 🧐
- Variant Analysis / Microbial Variant Calling 🧐
- Variant Analysis / Calling very rare variants 🧐
- Evolution / Phylogenetic analysis for bacterial comparative genomics 🧐
- Statistics and machine learning / Introduction to Machine Learning using R 🧐
- Single Cell / Pre-processing of Single-Cell RNA Data 🧐
- Single Cell / Single-cell quality control with scater 🧐
- Single Cell / Clustering 3K PBMCs with Scanpy 🧐
- Single Cell / Downstream Single-cell RNA analysis with RaceID 🧐
- Proteomics / Proteogenomics 1: Database Creation 🧐
- Galaxy Server administration / Automation with Jenkins 🧐
- Galaxy Server administration / Use Apptainer containers for running Galaxy jobs 🧐
- Galaxy Server administration / Galaxy Installation with Ansible 📝
- Galaxy Server administration / Galaxy Monitoring with Reports 🧐
- Galaxy Server administration / Ansible 🧐
- Galaxy Server administration / Distributed Object Storage 🧐
- Galaxy Server administration / Performant Uploads with TUS 🧐
- Galaxy Server administration / Galaxy Interactive Tools ✍️
- Galaxy Server administration / Galaxy Tool Management with Ephemeris 🧐
- Galaxy Server administration / Data Libraries 🧐
- Galaxy Server administration / Deploying a compute cluster in OpenStack via Terraform 🧐
Slides
- Sequence analysis / Quality Control ✍️ 🧐
- Sequence analysis / Mapping 🧐
- Transcriptomics / Integrate and query local datasets and distant RDF data with AskOmics using Semantic Web technologies ✍️ 🧐
- Genome Annotation / Introduction to Genome Annotation ✍️ 🧐
- Genome Annotation / Refining Genome Annotations with Apollo ✍️ 🧐
- Development in Galaxy / Prerequisites for building software/conda packages ✍️ 🧐
- Development in Galaxy / Galaxy Code Architecture 🧐
- Development in Galaxy / Tool development and integration into Galaxy ✍️ 🧐
- Development in Galaxy / Tool Shed: sharing Galaxy tools ✍️ 🧐
- Development in Galaxy / Tool Dependencies and Conda 🧐
- Assembly / Deeper look into Genome Assembly algorithms ✍️
- Assembly / Genome assembly and assembly QC - Introduction short version 📝 🧐
- Assembly / An introduction to get started in genome assembly and annotation 📝 🧐
- Assembly / Genome assembly quality control. ✍️ 🧐
- Introduction to Galaxy Analyses / Options for using Galaxy ✍️
- Introduction to Galaxy Analyses / Introduction to Galaxy 🧐
- Visualisation / JBrowse 🧐
- Visualisation / Visualisations in Galaxy 🧐
- Visualisation / Circos 🧐
- Galaxy Server administration / Server: Other 🧐
- Galaxy Server administration / Ansible 🧐
- Galaxy Server administration / Galaxy Interactive Tools ✍️ 🧐
- Galaxy Server administration / Storage Management 🧐
Video Recordings
- Genome Annotation / Genome annotation with Prokka 💬
- Genome Annotation / Refining Genome Annotations with Apollo 💬
- Genome Annotation / Long non-coding RNAs (lncRNAs) annotation with FEELnc 💬
- Genome Annotation / Genome annotation with Funannotate 💬 🗣
- Genome Annotation / Genome annotation with Prokka 💬 🗣
- Genome Annotation / Refining Genome Annotations with Apollo (prokaryotes) 💬 🗣
- Genome Annotation / Masking repeats with RepeatMasker 💬 🗣
- Galaxy Server administration / Galaxy Interactive Tools 💬 🗣
Events
GitHub Activity
github Issues Reported
67 Merged Pull Requests
See all of the github Pull Requests and github Commits by Anthony Bretaudeau.
-
 Update Jbrowse parameter
variant-analysisassemblygenome-annotation
Update Jbrowse parameter
variant-analysisassemblygenome-annotation -
 Fix input data for apollo tutorial (fixes #5792)
genome-annotation
Fix input data for apollo tutorial (fixes #5792)
genome-annotation -
 Update abretaud infos
template-and-tools
Update abretaud infos
template-and-tools -
 Add eurosciencegateway affiliation to some GGA stuff
template-and-toolsgenome-annotation
Add eurosciencegateway affiliation to some GGA stuff
template-and-toolsgenome-annotation -
 Annotation learning paths
template-and-toolsgenome-annotation
Annotation learning paths
template-and-toolsgenome-annotation
Reviewed 71 PRs
We love our community reviewing each other's work!
-
 add training comparison annotation
genome-annotation
add training comparison annotation
genome-annotation -
 Update training assembly
template-and-toolsassembly
Update training assembly
template-and-toolsassembly -
 Update tuto assembly pacbio hifi
assembly
Update tuto assembly pacbio hifi
assembly -
 Updated slides on short assembly/QC training to include srobin feedback
assembly
Updated slides on short assembly/QC training to include srobin feedback
assembly -
 typo error
genome-annotation
typo error
genome-annotation
News
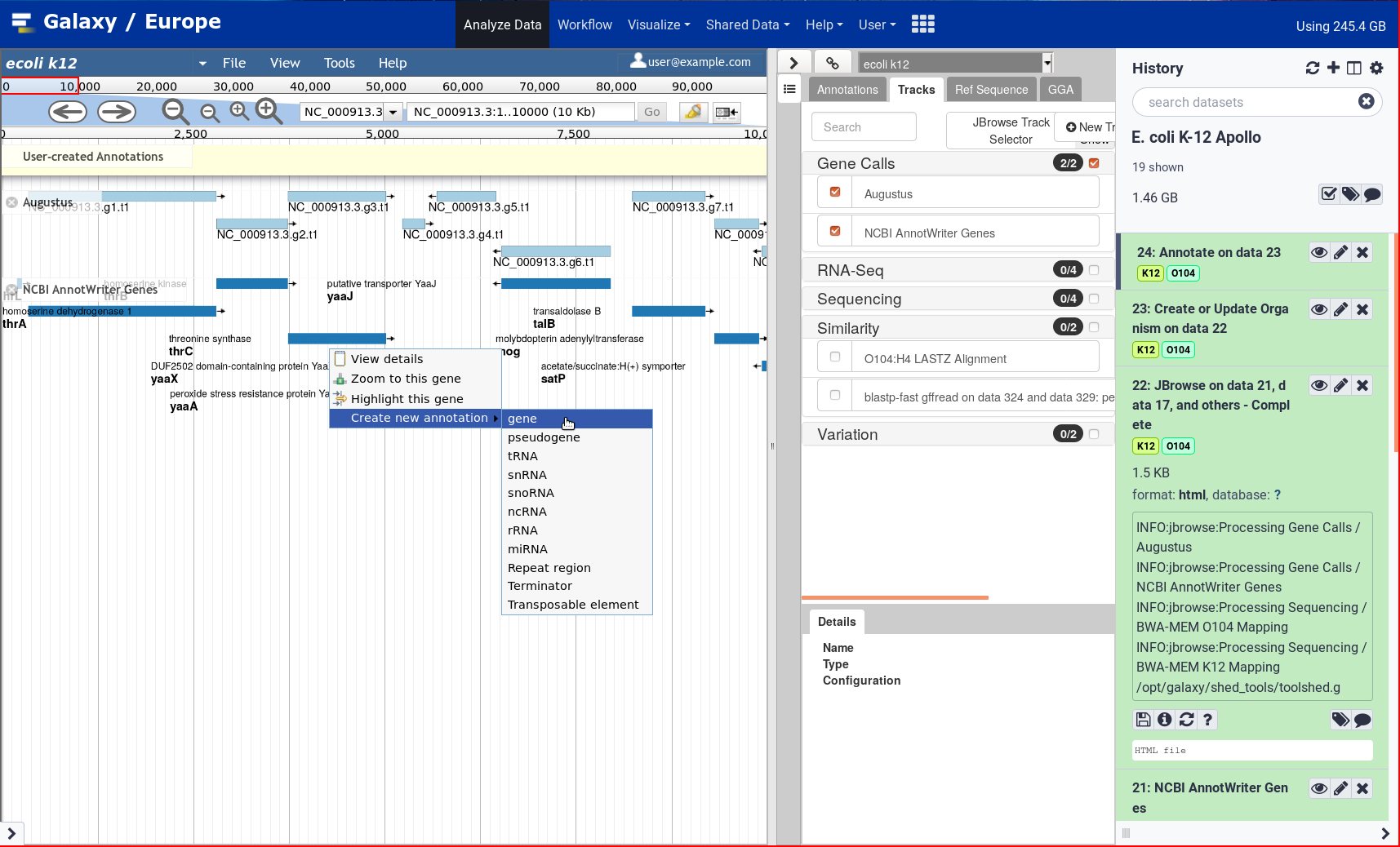
New Tutorial: Genome Annotation with Apollo
4 June 2021
After two years, the Galaxy Genome Annotation team has finally finished their much awaited tutorial on doing Genome Annotation in Galaxy! Genome annotation is a time consuming and largely manual process as it requires the synthesis of a significant amount of varied information to make inferences, a process tha cannot be accomplished without an interactive editor. To support this, Apollo was deployed on UseGalaxy.eu. Over the past two years the GGA project has pushed forward apollo and tooling around it, and it finally culminates in this look at how to annotated your genomes in Apollo.
External Links

Favourite Topics
Favourite Formats