Wolfgang Maier
Affiliations
Contributions
The following list includes only slides and tutorials where the individual or organisation has been added to the contributor list. This may not include the sum total of their contributions to the training materials (e.g. GTN css or design, tutorial datasets, workflow development, etc.) unless described by a news post.
Editorial Roles
This contributor has taken on additional responsibilities as an editor for the following topics. They are responsible for ensuring that the content is up to date, accurate, and follows GTN best practices.
- Topic: SARS-CoV-2
- Topic: One Health
- Topic: Variant Analysis
- Learning Pathway: Epigenetics data analysis with Galaxy
Tutorials
- Proteomics / MaxQuant and MSstats for the analysis of TMT data 🧐
- Ecology / RAD-Seq Reference-based data analysis 🧐
- Ecology / Data submission using ENA upload Tool 🧐
- Ecology / RAD-Seq de-novo data analysis 🧐
- Epigenetics / CUT&RUN data analysis 📝 🧐
- Epigenetics / DNA Methylation data analysis 📝 🧐
- Using Galaxy and Managing your Data / Automating Galaxy workflows using the command line ✍️ 🧐
- Transcriptomics / Genome-wide alternative splicing analysis 🧐
- Transcriptomics / Reference-based RNA-Seq data analysis 🧐
- Single Cell / Pre-processing of Single-Cell RNA Data ✍️
- Variant Analysis / Avian influenza viral strain analysis from gene segment sequencing data ✍️ 🧐
- Variant Analysis / Exome sequencing data analysis for diagnosing a genetic disease ✍️ 🧐
- Variant Analysis / Mutation calling, viral genome reconstruction and lineage/clade assignment from SARS-CoV-2 sequencing data ✍️ 🧐
- Variant Analysis / Mapping and molecular identification of phenotype-causing mutations ✍️ 🧐
- Variant Analysis / From NCBI's Sequence Read Archive (SRA) to Galaxy: SARS-CoV-2 variant analysis 📝
- Variant Analysis / Deciphering Virus Populations - Single Nucleotide Variants (SNVs) and Specificities in Baculovirus Isolates 🧐
- Variant Analysis / Pox virus genome analysis from tiled-amplicon sequencing data ✍️ 🧐
- Variant Analysis / Identification of somatic and germline variants from tumor and normal sample pairs ✍️ 🧐
- Variant Analysis / Somatic Variant Discovery from WES Data Using Control-FREEC ✍️ 🧐
- Variant Analysis / Trio Analysis using Synthetic Datasets from RD-Connect GPAP ✍️ 🧐
- Variant Analysis / M. tuberculosis Variant Analysis 🧐
- Galaxy Server administration / Data Libraries 🧐
- Galaxy Server administration / Galaxy Tool Management with Ephemeris 🧐
- Contributing to the Galaxy Training Material / Creating a new tutorial 🧐
- Microbiome / Pathogen detection from (direct Nanopore) sequencing data using Galaxy - Foodborne Edition 📝 🧐
- Introduction to Galaxy Analyses / Galaxy Basics for genomics 🧐
- Sequence analysis / Primer and primer scheme design for pan-specific detection and sequencing of viral pathogens across genotypes ✍️ 🧐
- Sequence analysis / Quality and contamination control in bacterial isolate using Illumina MiSeq Data 🧐
- Sequence analysis / Quality Control 🧐
- Sequence analysis / Removal of human reads from SARS-CoV-2 sequencing data ✍️ 🧐
- Assembly / Unicycler assembly of SARS-CoV-2 genome with preprocessing to remove human genome reads ✍️ 🧐
- Assembly / Genome Assembly of a bacterial genome (MRSA) sequenced using Illumina MiSeq Data 🧐
- Evolution / Identifying tuberculosis transmission links: from SNPs to transmission clusters 📝 🧐
- Evolution / Tree thinking for tuberculosis evolution and epidemiology 📝 🧐
Slides
FAQs
- How do I create an account on a public Galaxy instance?
- Is it possible to use alternative tools to those proposed in the tutorial?
- Pick the right Concatenate tool
- Adding a tag
- Creating a new file
- Importing data from a data library
- Importing data from remote files
- Renaming a dataset
- Créer un nouvel history
- Creating a new history
- Renaming a history
- Select multiple datasets
- Downloading the files from the NCBI server fails or takes too long.
- Opening the workflow editor
- Importing a workflow
- Importing a workflow using the Tool Registry Server (TRS) search
- Make a workflow public
- Renaming workflow outputs
- Running a workflow
- Importing and Launching a Dockstore Workflow
- Importing and Launching a WorkflowHub.eu Workflow
Video Recordings
- Variant Analysis / Mutation calling, viral genome reconstruction and lineage/clade assignment from SARS-CoV-2 sequencing data 💬 🗣
Events
- Workshop on high-throughput sequencing data analysis with Galaxy 🧑🏫
- Workshop on high-throughput sequencing data analysis with Galaxy 🧑🏫
- Galaxy Training Academy 2025 🧑🏫
- Galaxy Training Academy 2024 🧑🏫
GitHub Activity
github Issues Reported
63 Merged Pull Requests
See all of the github Pull Requests and github Commits by Wolfgang Maier.
-
 Update CONTRIBUTORS.yaml
template-and-tools
Update CONTRIBUTORS.yaml
template-and-tools -
 Add new learning pathway for epigenetics
template-and-tools
Add new learning pathway for epigenetics
template-and-tools -
 Update to latest cutadapt interface in quality control
transcriptomicssequence-analysis
Update to latest cutadapt interface in quality control
transcriptomicssequence-analysis -
 Add link to AIV reference history on Galaxy US
variant-analysis
Add link to AIV reference history on Galaxy US
variant-analysis -
 BY-COVID track update
template-and-tools
BY-COVID track update
template-and-tools
Reviewed 44 PRs
We love our community reviewing each other's work!
-
 Change from fastqc to falco in somatic-variant-discovery tutorial
variant-analysis
Change from fastqc to falco in somatic-variant-discovery tutorial
variant-analysis -
 Change from FastQC to Falco in the tutorials quality-contamination-control and mrsa-illumina
assemblysequence-analysis
Change from FastQC to Falco in the tutorials quality-contamination-control and mrsa-illumina
assemblysequence-analysis -
 Add myself as contributor in ref-based-rad-seq tutorial
ecology
Add myself as contributor in ref-based-rad-seq tutorial
ecology -
 Change to latest MAFFT version in viral-primer-design tutorial
sequence-analysis
Change to latest MAFFT version in viral-primer-design tutorial
sequence-analysis -
 Change from fastqc to falco in somatic-variants tutorial
variant-analysis
Change from fastqc to falco in somatic-variants tutorial
variant-analysis
News
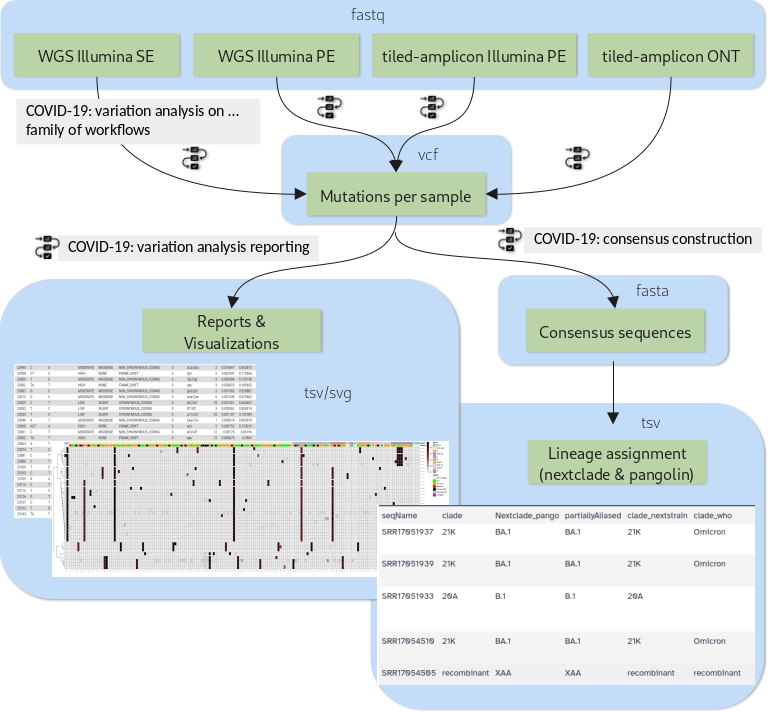
New Tutorial: Mutation calling, viral genome reconstruction and lineage/clade assignment from SARS-CoV-2 sequencing data
30 June 2021
Effectively monitoring global infectious disease crises, such as the COVID-19 pandemic, requires capacity to generate and analyze large volumes of sequencing data in near real time. These data have proven essential for monitoring the emergence and spread of new variants, and for understanding the evolutionary dynamics of the virus.
New GTN Feature Tag-based Topics enables new SARS-CoV-2 topic
23 January 2023
The GTN has long struggled with the rigid hierarchy of a single level of classification: topics. All materials had to be organised into a single topic, even when their contents sometimes spanned multiple topics! We added subtopics a while back to help further organise these tutorials, but that still did not solve the issue of needing to collect multiple learn materials in a single location for people to access.
External Links

Favourite Topics
Favourite Formats