18 November 2022
new topic
single-cell
new feature
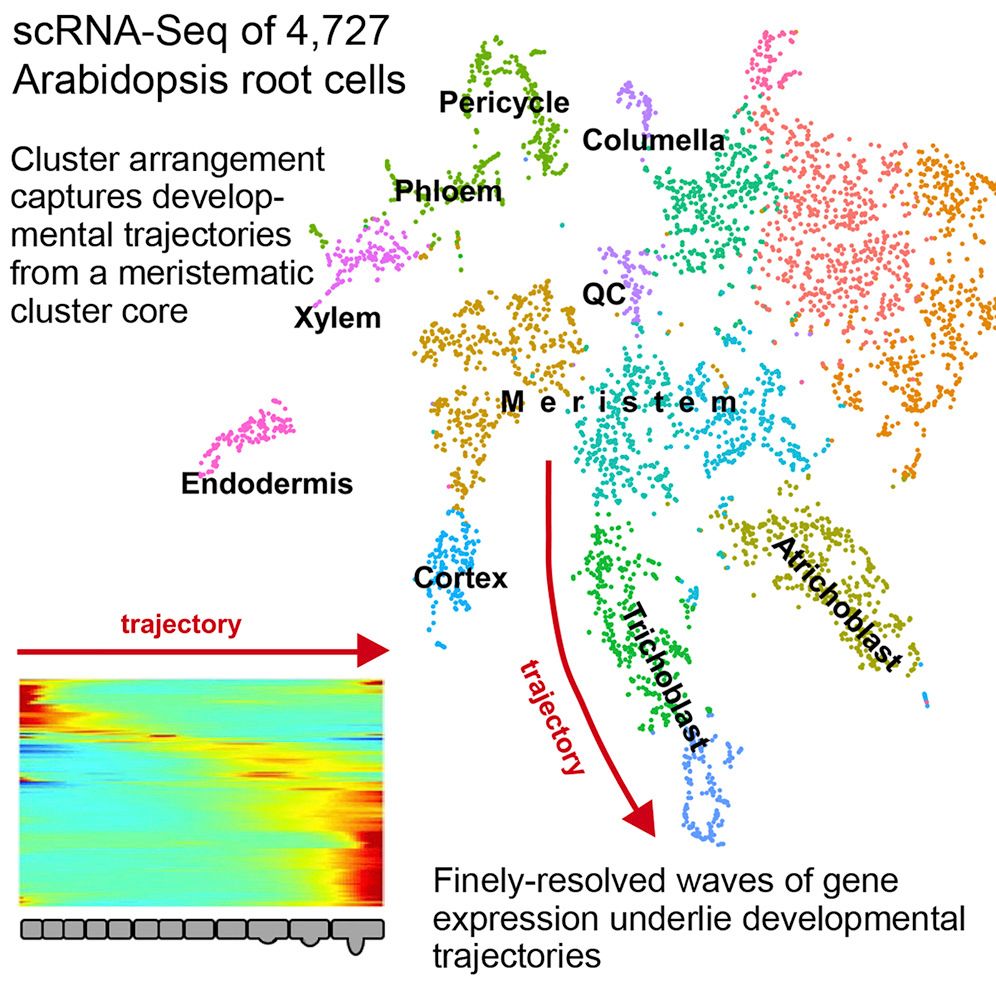
Single-cell analysis now has it’s own topic! These tutorials were previously part of the transcriptomics topic, but due to the amazing efforts by
Wendi Bacon,
Mehmet Tekmanand others, we now have so many single-cell analysis tutorials that they deserve their own dedicated topic! From introductory slides and practicals, to case study tutorials generating cell clusters and trajectories from raw sequencing files, and even a growing subtopic of smaller tips and tricks for adapting analysis to user needs, the single cell topic either has what you need or is working on it. Come learn or get involved!
Full Story