| Author(s) |
|
| Editor(s) |
|
| Infrastructure |
|
Posted on: 31 January 2024 purlPURL: https://gxy.io/GTN:N00069
Hey Galaxy admins! Have you ever wanted to administer your own instance from the luxury of your pocket?
Now you can, with Galaxy In Your Pocket™!
Why?
Run your instance without having to worry about the usual concerns: a noisy server, too many cables, bad internet, or just a plain unwillingness to spend time away from a screen.
Having Galaxy In Your Pocket™, you can run and administer your own instance all from the same portable device that you use to ignore your loved ones with!
How?
With just a few steps, you can have Galaxy running on a mobile ARM device of your own.
 Open image in new tab
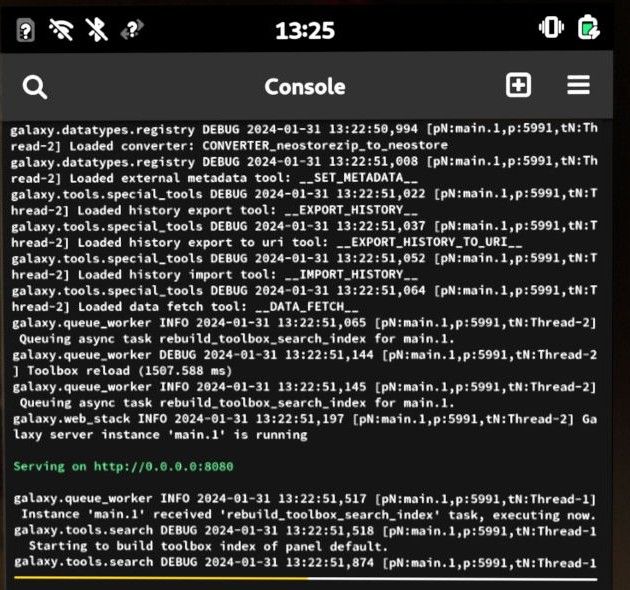
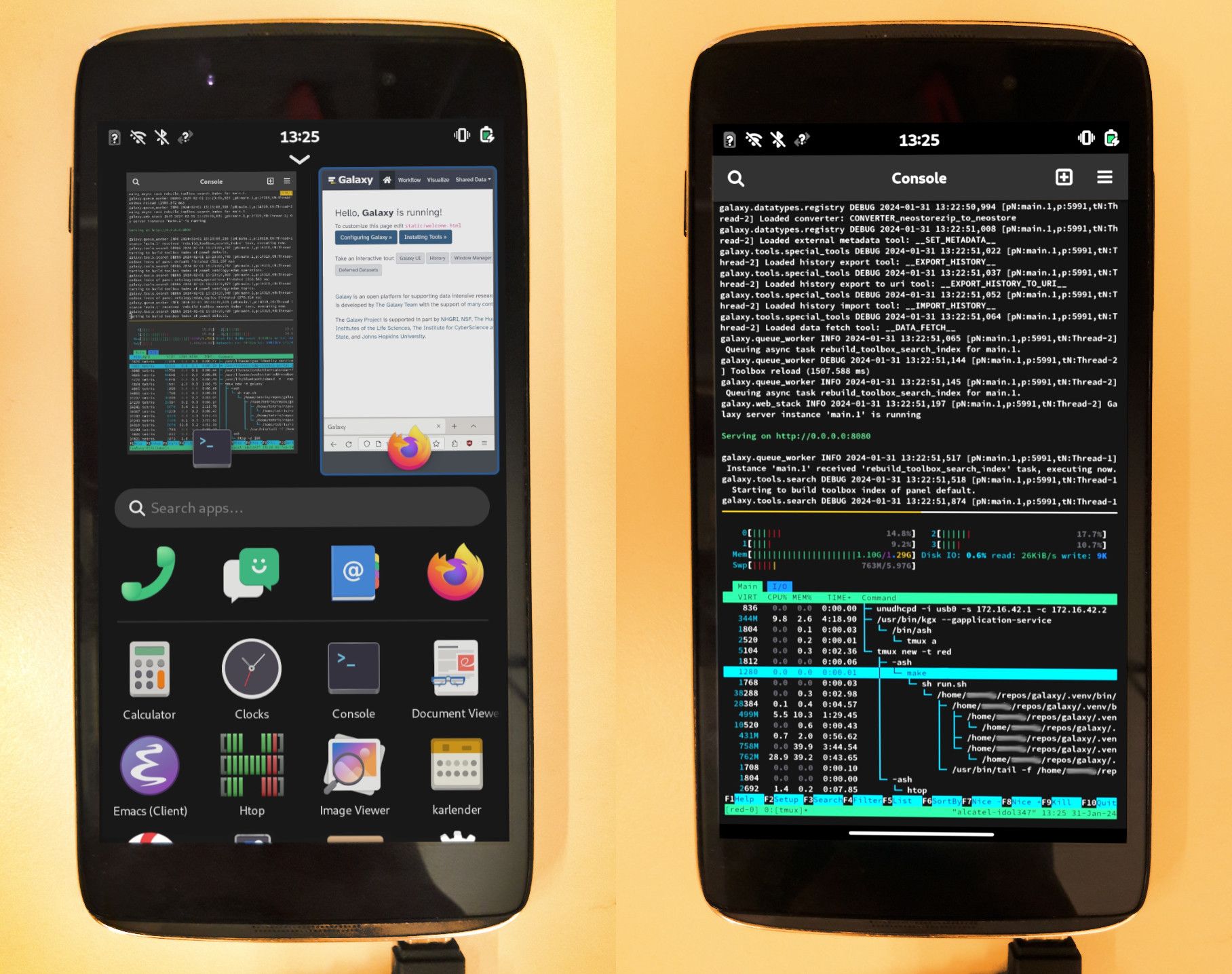
Open image in new tabAbove is an image of Galaxy running within Phosh UI in PostmarketOS on an old Alcatel Idol 347, using a Quad-core 1.2 GHz Cortex-A53 CPU and 1.3GB of RAM, with a 5GB swap mounted.
Galaxy appears to use around ~1 GB RAM, and the phone still has enough memory to spare to access the Galaxy UI via Firefox.
Really, why?
Just imagine: Galaxy In Your Pocket™! A united Galaxy Client and Galaxy Server running on your phone 📲, which you can then access using that same phone’s web browser.
It’s the perfect setup that only a true and dedicated Galaxy Admin such as yourself can dream about.
Or if you’re not buying any of this, then it’s for those emergency situations where you forgot your laptop and need to solve a bioinformatics crisis on the go 🧬🌄