Alex Ostrovsky
Contributions
The following list includes only slides and tutorials where the individual or organisation has been added to the contributor list. This may not include the sum total of their contributions to the training materials (e.g. GTN css or design, tutorial datasets, workflow development, etc.) unless described by a news post.
Tutorials
- Assembly / Vertebrate genome assembly using HiFi, Bionano and Hi-C data - Step by Step ✍️
- Assembly / Using the VGP workflows to assemble a vertebrate genome with HiFi and Hi-C data ✍️
- Variant Analysis / Calling variants in non-diploid systems ✍️ 🧐
- Single Cell / Downstream Single-cell RNA analysis with RaceID ✍️
- Introduction to Galaxy Analyses / Galaxy Basics for everyone 🧐
- Development in Galaxy / Creating Galaxy tools from Conda Through Deployment ✍️ 🧐
Slides
- Single Cell / Plates, Batches, and Barcodes ✍️
- Single Cell / Dealing with Cross-Contamination in Fixed Barcode Protocols ✍️
- Development in Galaxy / Tool Dependencies and Containers 🧐
- Development in Galaxy / Tool Dependencies and Conda 🧐
FAQs
- The folder `recipes/belerophon/` and the file `meta.yaml` already exist in bioconda?
- Where can I get planemo?
Video Recordings
- Introduction to Galaxy Analyses / Galaxy Basics for everyone 💬 🗣
- Development in Galaxy / Creating Galaxy tools from Conda Through Deployment 💬 🗣
GitHub Activity
github Issues Reported
8 Merged Pull Requests
See all of the github Pull Requests and github Commits by Alex Ostrovsky.
-
 Add my orcid
template-and-tools
Add my orcid
template-and-tools -
 Add tools from scratch training
Add tools from scratch training
-
 add vgp training skeleton from workflow (hidden)
add vgp training skeleton from workflow (hidden)
-
 workflows updated with newer tools, added missing
workflows updated with newer tools, added missing
-
 Data link modernized
Data link modernized
Reviewed 2 PRs
We love our community reviewing each other's work!
News
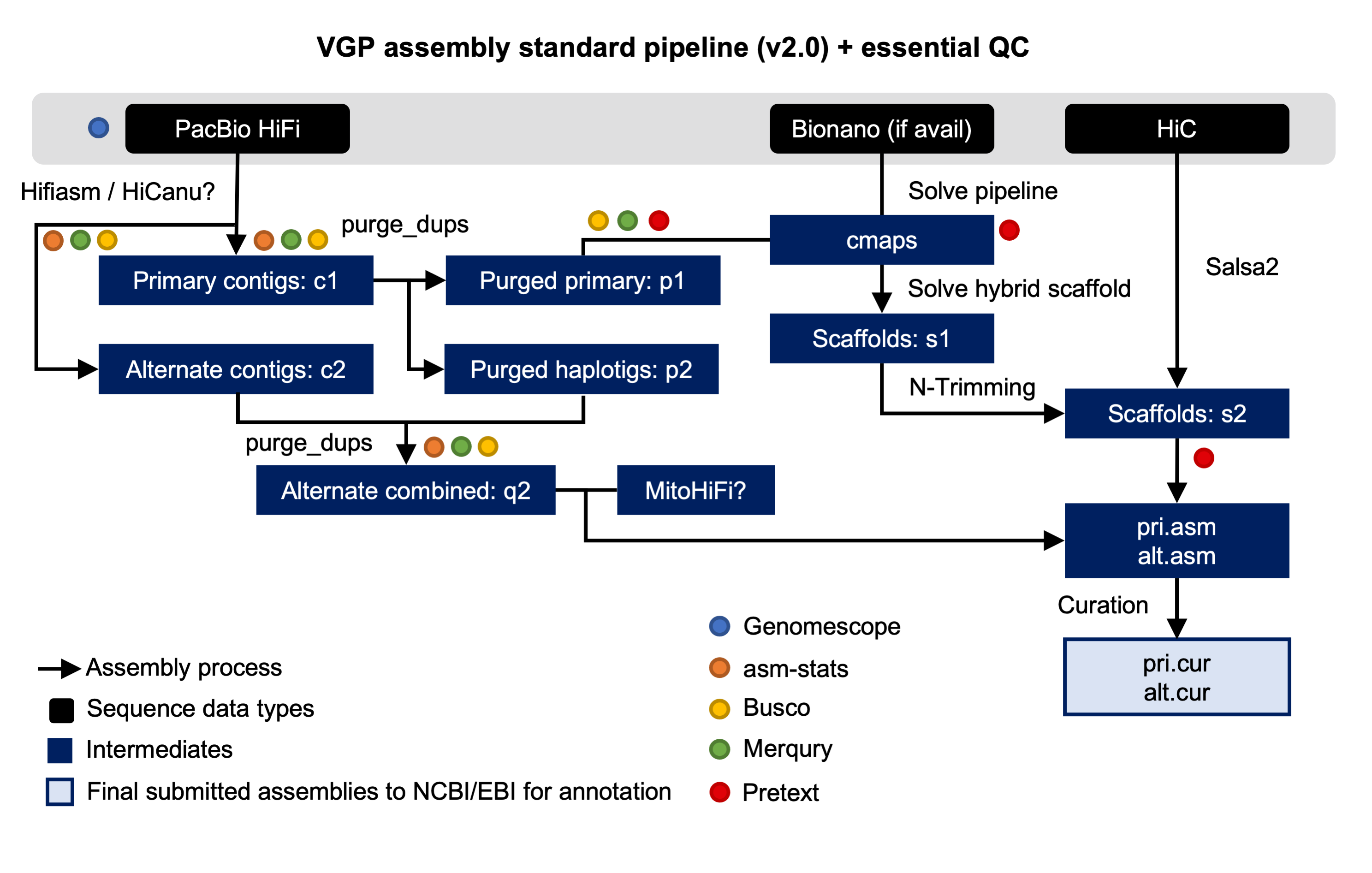
New Tutorial: VGP assembly pipeline
14 March 2022
We are proud to announce that, as result of the collaboration with the Vertebrate Genomes Project (VGP), a new training describing the VGP assembly pipeline is now available in the Galaxy Training Network. The Vertebrate Genomes Project aims to generate high-quality, near-error-free, gap-free, chromosome-level, haplotype-phased, annotated reference genome assemblies for every vertebrate species.
External Links
Favourite Topics
Favourite Formats