Lucille Delisle
Affiliations
Contributions
The following list includes only slides and tutorials where the individual or organisation has been added to the contributor list. This may not include the sum total of their contributions to the training materials (e.g. GTN css or design, tutorial datasets, workflow development, etc.) unless described by a news post.
Editorial Roles
This contributor has taken on additional responsibilities as an editor for the following topics. They are responsible for ensuring that the content is up to date, accurate, and follows GTN best practices.
- Topic: Epigenetics
Tutorials
- Proteomics / metaQuantome 1: Data creation 🧐
- Epigenetics / CUT&RUN data analysis 📝 🧐
- Epigenetics / ATAC-Seq data analysis ✍️ 🧐
- Using Galaxy and Managing your Data / Group tags for complex experimental designs 🧐
- Using Galaxy and Managing your Data / Understanding Galaxy history system 🧐
- Transcriptomics / Genome-wide alternative splicing analysis ✍️ 🧐
- Transcriptomics / Network analysis with Heinz 🧐
- Transcriptomics / Reference-based RNA-Seq data analysis ✍️ 🧐
- Transcriptomics / RNA-seq Alignment with STAR 🧐
- Transcriptomics / CLIP-Seq data analysis from pre-processing to motif detection 🧐
- Single Cell / Understanding Barcodes 🧐
- Variant Analysis / Trio Analysis using Synthetic Datasets from RD-Connect GPAP 🧐
- Galaxy Server administration / External Authentication 🧐
- Galaxy Server administration / Connecting Galaxy to a compute cluster 🧐
- Galaxy Server administration / Galaxy Monitoring with Reports 🧐
- Galaxy Server administration / Reference Data with CVMFS 🧐
- Galaxy Server administration / Server Maintenance: Cleanup, Backup, and Restoration ✍️ 🧐
- Galaxy Server administration / Reference Data with CVMFS without Ansible 🧐
- Galaxy Server administration / Ansible 🧐
- Galaxy Server administration / Running Jobs on Remote Resources with Pulsar 🧐
- Galaxy Server administration / Enable upload via FTP ✍️ 🧐
- Galaxy Server administration / Use Apptainer containers for running Galaxy jobs 🧐
- Galaxy Server administration / Data Libraries 🧐
- Galaxy Server administration / Mapping Jobs to Destinations using TPV 🧐
- Galaxy Server administration / Distributed Object Storage 🧐
- Galaxy Server administration / Automation with Jenkins 🧐
- Galaxy Server administration / Galaxy Monitoring with gxadmin 🧐
- Galaxy Server administration / Galaxy Monitoring with Telegraf and Grafana 🧐
- Galaxy Server administration / Galaxy Tool Management with Ephemeris 🧐
- Galaxy Server administration / Deploying Wireguard for private mesh networking 🧐
- Galaxy Server administration / Training Infrastructure as a Service (TIaaS) 🧐
- Galaxy Server administration / Performant Uploads with TUS ✍️ 🧐
- Galaxy Server administration / Galaxy Installation with Ansible 📝 🧐
- Galaxy Server administration / Galaxy Interactive Tools 🧐
- Contributing to the Galaxy Training Material / Creating content in Markdown 🧐
- Contributing to the Galaxy Training Material / Creating a new tutorial ✍️
- Contributing to the Galaxy Training Material / Updating diffs in admin training 🧐
- Introduction to Galaxy Analyses / How to reproduce published Galaxy analyses 🧐
- Evolution / Identifying tuberculosis transmission links: from SNPs to transmission clusters 🧐
Slides
- Epigenetics / Introduction to ATAC-Seq data analysis ✍️
- Epigenetics / Introduction to DNA Methylation data analysis 🧐
- Transcriptomics / Identification of non-canonical ORFs and their potential biological function 📝 🧐
- Galaxy Server administration / Server Maintenance: Cleanup, Backup, and Restoration 🧐
- Galaxy Server administration / Controlling Galaxy with systemd or Supervisor 🧐
- Galaxy Server administration / Galaxy Administrator Time Burden and Technology Usage 🖥
- Galaxy Server administration / Galaxy Installation with Ansible 🧐
FAQs
- Automatically trim adapters (without providing custom sequences)
- In bowtie 2 parameters, in place of 1000 for other experiments, should we mention the median fragment length observed in our library?
- Creating a paired collection
- Changing the datatype
- Converting the file format
- Updating from 22.01 to 23.0 with Ansible
- Changing the heatmap colours
- Copy a dataset between histories
- Is it possible to subsample some samples if you have more reads?
- UCSC import: what should my file look like?
Video Recordings
- Epigenetics / Introduction to ATAC-Seq data analysis 💬 🗣
- Transcriptomics / Reference-based RNA-Seq data analysis 💬 🗣
Events
GitHub Activity
github Issues Reported
38 Merged Pull Requests
See all of the github Pull Requests and github Commits by Lucille Delisle.
-
 CUT&RUN: Fix zenodo link (left from ATAC copy paste)
epigenetics
CUT&RUN: Fix zenodo link (left from ATAC copy paste)
epigenetics -
 Update CUT&RUN tutorial
epigenetics
Update CUT&RUN tutorial
epigenetics -
 update training for CYOA if space in option
contributing
update training for CYOA if space in option
contributing -
 Fix CYOA of collection split in Ref-based
transcriptomics
Fix CYOA of collection split in Ref-based
transcriptomics -
 Add a third way to run DESeq2 in Ref-based tuto
transcriptomics
Add a third way to run DESeq2 in Ref-based tuto
transcriptomics
Reviewed 29 PRs
We love our community reviewing each other's work!
-
 Update CONTRIBUTORS.yaml
template-and-tools
Update CONTRIBUTORS.yaml
template-and-tools -
 Update CUT and RUN tutorial
epigenetics
Update CUT and RUN tutorial
epigenetics -
 Update CUT&RUN tutorial
epigenetics
Update CUT&RUN tutorial
epigenetics -
 Add a third way to run DESeq2 in Ref-based tuto
transcriptomics
Add a third way to run DESeq2 in Ref-based tuto
transcriptomics -
 Update FAQ about 22.01 to 23.0
admin
Update FAQ about 22.01 to 23.0
admin
News
New Feature: Trainer Directory! (Add yourself today!)
21 March 2023
Numerous other training groups keep “Trainer Directories” as a way to help students meet trainers in their area, especially for hosting workshops for their local community. There have been a handful of attempts at this in the past, both in galaxyproject/galaxy-maps and in a Google Map on the GTN homepage.
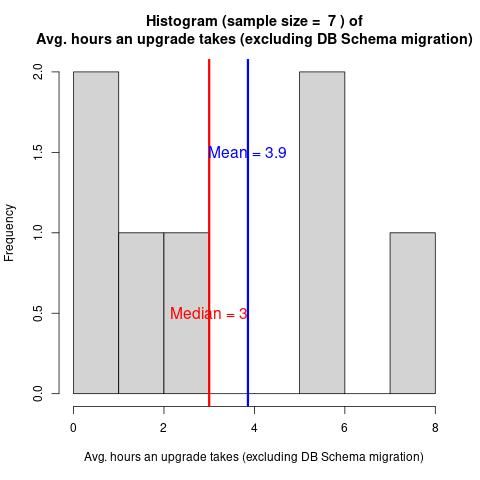
Galaxy Administrator Time Burden and Technology Usage
22 July 2024
Have you wondered how difficult Galaxy is to run? How much time people must spend to run Galaxy?
In February 2024, we collected 9 responses from the Galaxy Small Scale Admin group.
The questions cover various time burdens and technological choices.
The report provides answers to prospective future admins’ most common questions.
External Links

Favourite Topics
Favourite Formats