Lorum Ipsum
| Auteur·rice·s |
|
| Fournisseur·euse·s de données |
|
| Traduction |
|
| Éditeur·rice·s |
|
| Testeur·euse·s |
|
| Réviseur·euse·s |
|
| Expérience utilisateur·rice·s/Designer·euse·s |
|
| Infrastructure |
|
Description généraleQuestions:
Objectifs:
Lorem ipsum dolor sit amet,
onsectetur adipiscing elit.
Pré-requis:
Vestibulum sit amet velit nibh
Vivamus tincidunt diam ligula, id rutrum purus rutrum a.
Durée estimée: 42 MinutesNiveau: Introduction Introductorysupports:
Publication: Feb 19, 2025Dernière modification: Feb 19, 2025Licence: Le contenu de ce didacticiel est sous licence Creative Commons Attribution 4.0 International License. Le Framework GTN est sous licence MITversion Révision: 1
Test Tutorial for Language: fr
Lorem ipsum dolor sit amet, consectetur adipiscing elit. Vestibulum sit amet velit nibh. Nullam porttitor urna at mi vulputate convallis. Cras mattis tempor interdum.
Test all boxes
En pratique: Lorem Ipsum
- Lorem Ipsum ..
- Lorem Ipsum ..
Commentaire: Lorem Ipsum
- Lorem Ipsum ..
Question: Lorem Ipsum
- Lorem Ipsum ..
- Lorem Ipsum ..
- Lorem Ipsum ..
Attention: Lorem Ipsum
- Lorem Ipsum ..
Entrée: Lorem Ipsum
- Lorem Ipsum ..
Sortie: Lorem Ipsum
- Lorem Ipsum ..
Test snippets of different box types
To create a new history simply click the new-history icon at the top of the history panel:
En pratique: Importing and Launching a Dockstore Workflow
- Go to galaxy-workflows-activity Workflows → Import in your Galaxy
- Switch tabs to TRS ID
- Ensure the “TRS server” is set to “Dockstore”
- Provide your “TRS ID” (copied from your workflow’s Dockstore page)
- Select the workflow version you want to import
Commentaire: Nanopore sequencingNanopore sequencing has several properties that make it well-suited for our purposes
- Long-read sequencing technology offers simplified and less ambiguous genome assembly
- Long-read sequencing gives the ability to span repetitive genomic regions
- Long-read sequencing makes it possible to identify large structural variations
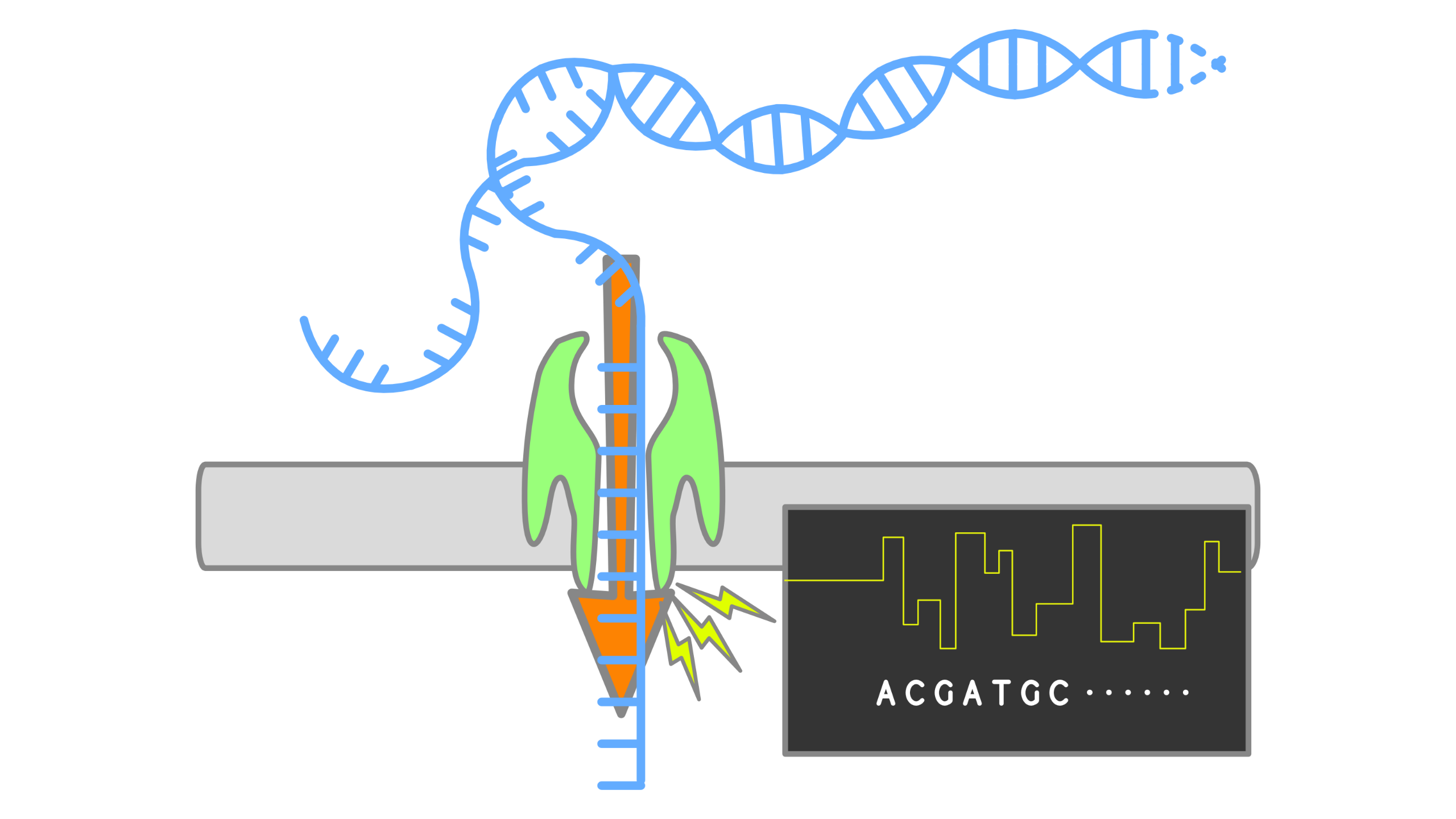
When using Oxford Nanopore Technologies (ONT) sequencing, the change in electrical current is measured over the membrane of a flow cell. When nucleotides pass the pores in the flow cell the current change is translated (basecalled) to nucleotides by a basecaller. A schematic overview is given in the picture above.
When sequencing using a MinIT or MinION Mk1C, the basecalling software is present on the devices. With basecalling the electrical signals are translated to bases (A,T,G,C) with a quality score per base. The sequenced DNA strand will be basecalled and this will form one read. Multiple reads will be stored in a fastq file.

