Avans Hogeschool
Members
These individuals have noted that they are affiliated in some way with this organisation. This list is non-exhaustive, and randomly ordered.
Former Members
These individuals have noted that they previously were affiliated in some way with this organisation. This list is non-exhaustive.
Contributions
The following list includes only slides and tutorials where the individual or organisation has been added to the contributor list. This may not include the sum total of their contributions to the training materials (e.g. GTN css or design, tutorial datasets, workflow development, etc.) unless described by a news post.
Tutorials
- Assembly / Genome Assembly of MRSA from Oxford Nanopore MinION data (and optionally Illumina data) 💵
- Assembly / Genome Assembly of a bacterial genome (MRSA) sequenced using Illumina MiSeq Data 💵
- Genome Annotation / Identification of AMR genes in an assembled bacterial genome 💵
- Foundations of Data Science / Python - Coding Style 💵
- Foundations of Data Science / Conda Environments For Software Development 💵
- Foundations of Data Science / SQL with Python ✍️
- Foundations of Data Science / SQL Educational Game - Murder Mystery 💵
- Foundations of Data Science / Advanced CLI in Galaxy ✍️
- Foundations of Data Science / Introduction to SQL ✍️
- Foundations of Data Science / CLI basics ✍️
- Foundations of Data Science / dplyr & tidyverse for data processing ✍️
- Foundations of Data Science / Python - Multiprocessing 💵
- Foundations of Data Science / Python - Argparse 💵
- Foundations of Data Science / Virtual Environments For Software Development 💵
- Foundations of Data Science / Make & Snakemake ✍️
- Foundations of Data Science / Python - Type annotations 💵
- Foundations of Data Science / Advanced SQL ✍️
- Foundations of Data Science / SQL with R ✍️
- Foundations of Data Science / Python - Testing 💵
Learning Pathways
GitHub Activity
github Issues Reported
News
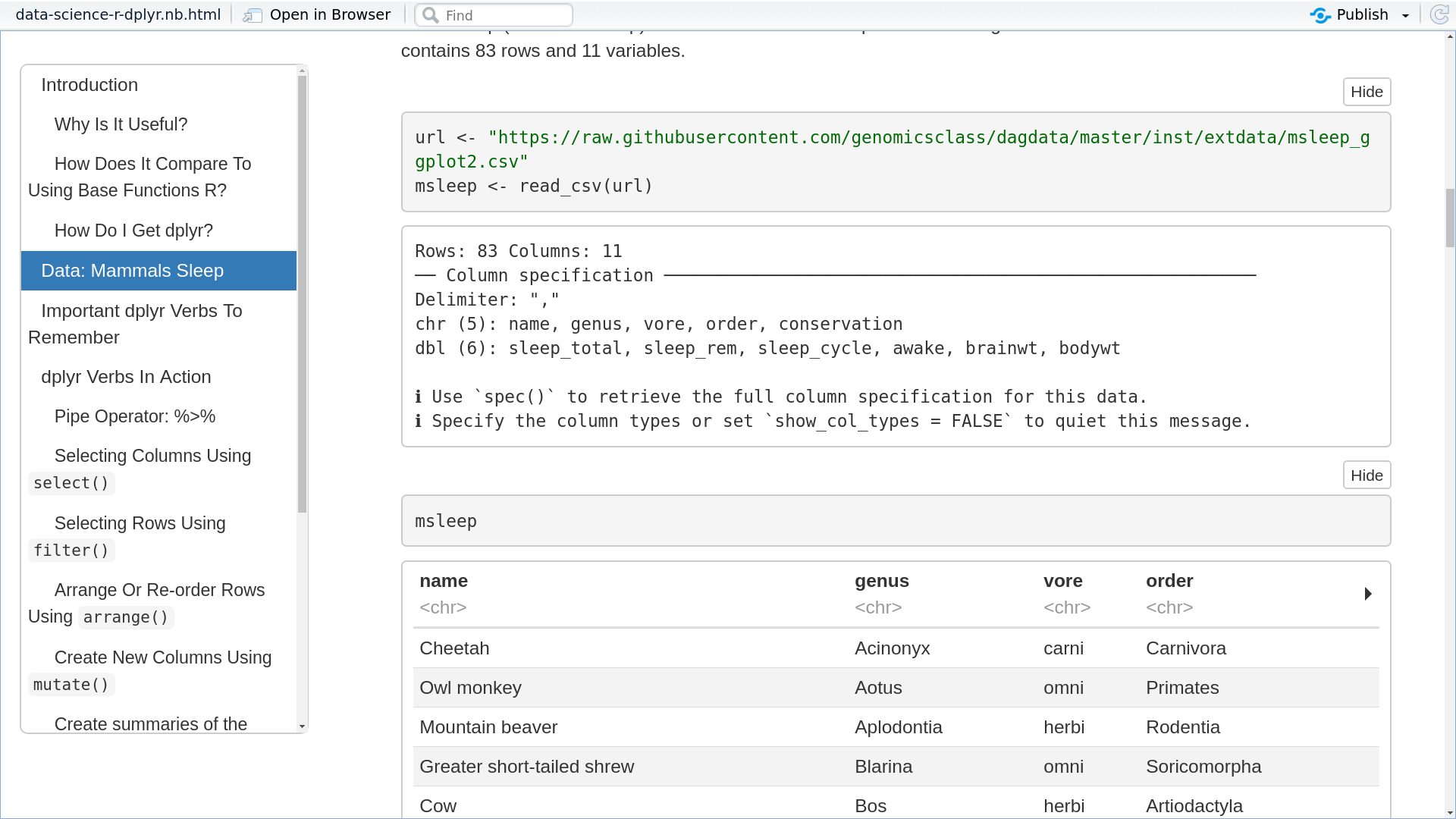
New Feature: Automatic RMarkdown
 28 January 2022
Further building on the work in the automatic Jupyter Notebook, we’ve now re-written the Jupyter export to be faster, and more importantly added support for R and RMarkdown! Check out an example material. Here we take the content of the tutorial, written like normal GTN markdown, and we automatically convert it to Jupyter Notebooks and now RMarkdown documents! Check out the documentation on how to setup your tutorials to support this.
28 January 2022
Further building on the work in the automatic Jupyter Notebook, we’ve now re-written the Jupyter export to be faster, and more importantly added support for R and RMarkdown! Check out an example material. Here we take the content of the tutorial, written like normal GTN markdown, and we automatically convert it to Jupyter Notebooks and now RMarkdown documents! Check out the documentation on how to setup your tutorials to support this.

New Feature: Automatic RMarkdown
28 January 2022
Further building on the work in the automatic Jupyter Notebook, we’ve now re-written the Jupyter export to be faster, and more importantly added support for R and RMarkdown! Check out an example material. Here we take the content of the tutorial, written like normal GTN markdown, and we automatically convert it to Jupyter Notebooks and now RMarkdown documents! Check out the documentation on how to setup your tutorials to support this.
