| Author(s) |
|
| Infrastructure |
|
Posted on: 13 May 2024 purlPURL: https://gxy.io/GTN:N00078
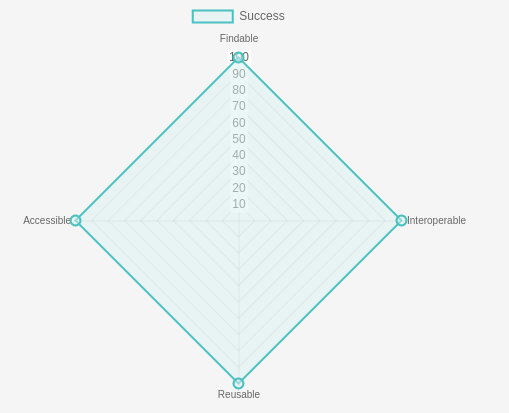
We are happy to report that over the years of our fruitful collaboration with ELIXIR TeSS, all GTN materials should receive a perfect score on the FAIR-Checker.
This achievement is a reflection of our long standing and ongoing commitment to ensuring all of the materials you share with others via the GTN are automatically FAIR: Findable, Accessible, Interoperable, and Reusable. Of course the GTN goes further meeting criteria for FAIR/O, as all of our resources are shared with the community under an Open License.
Why the GTN?
Amongst other competitors in the bioinformatics training material repository space the GTN is unique, we are the only one (which we’re aware of!) receiving a perfect score on the FAIR-Checker.
 Open image in new tab
Open image in new tabFor Tutorial Authors
We have made no new requirements for tutorial authors, simply by contributing a tutorial to the GTN which passes our linting, you can be sure it meets all relevant FAIR criteria due to our infrastructure and automation.
For Trainers
If you’re a trainer, you can be confident that all GTN materials you use in your training are FAIR. Please let your funders know you are meeting these criteria as well!
View Material