| Author(s) |
|
Posted on: 15 May 2023 purlPURL: https://gxy.io/GTN:N00053
The GTN hosts a new training for analyzing alternative splicing at genome-wide scale!
New Galaxy training: Genome-wide alternative splicing analysis
Isoform switching is a biological phenomenon that plays a crucial role in the regulation and diversity of gene expression. It refers to the process by which alternative splicing of pre-mRNA generates different mRNA isoforms that can produce distinct protein products under different conditions. Despite the fact that alternative splicing has been demonstrated to play a fundamental roles in a wide range of biological contexts, such as tissue-specific expression, disease development and evolutionary adaptation, isoform-based expression anaysis pipelines are still rare when compared with gene-level quantification studies.
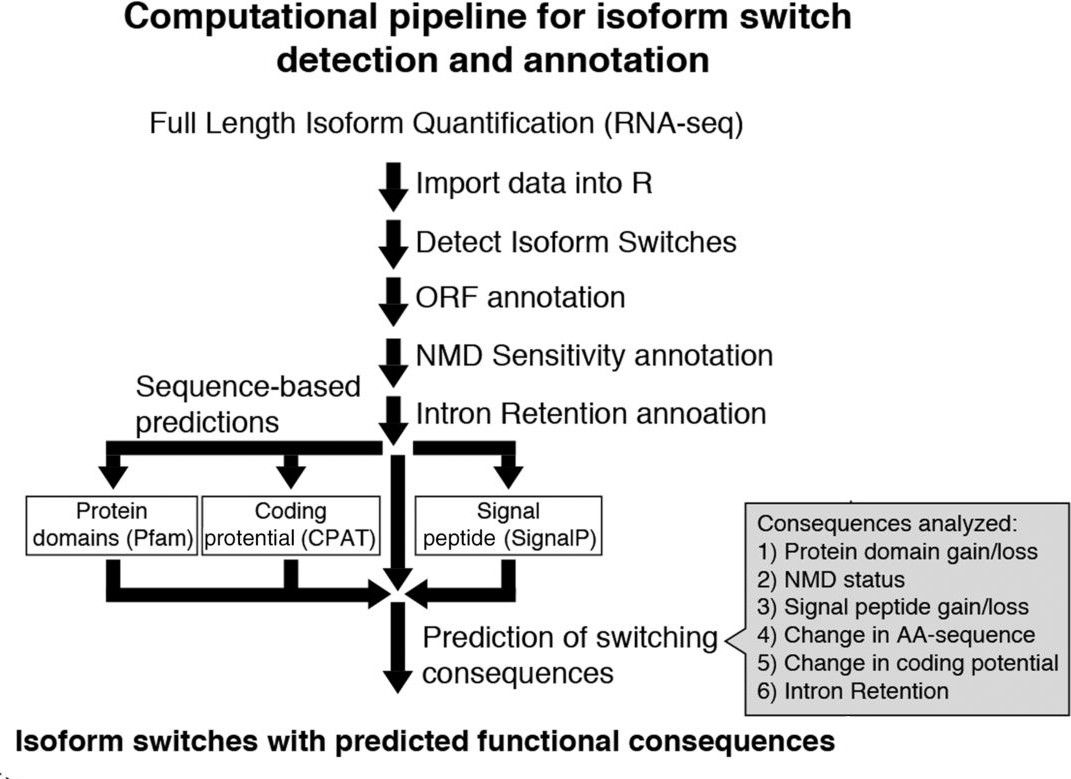
This new Galaxy training explores the isoform switching concept and provides a step-by-step guide for analyzing this regulatory mechanism. It makes use of IsoformSwitchAnalyzeR, a R package which integrates a multi-step analysis for isoform detection and functional prediction.
 Open image in new tab
Open image in new tabA second important aspect of this pipeline is that it allows to evaluate alternative splicing at two different levels: gene-specific and genome-wide scale. This dual-approach allows not only to indentify specific genes, but also prividing an analytical instrument for the study of genome-wide regulatory networks. Finally, a third important element of this training, directly derived from the operational capabilities of IsoformSwitchAnalyzeR, is that it allows to integrate multiple layers of information in the analysis. Concretely, in order to evaluate the functional consequences of the isoform switching events, IsoformSwitchAnalyzeR allows to integrate information bout coding-potential, protein-domains, signal peptides and instrinsically disordered regions.
View Material