| Author(s) |
|
Posted on: 26 January 2023 purlPURL: https://gxy.io/GTN:N00043
Food contamination with pathogens are a major burden on our society. Globally, they affect an estimated 600 million people a year and impact socioeconomic development at different levels. These outbreaks are mainly due to Salmonella spp. followed by Campylobacter spp. and Noroviruses.
The evolution of techniques in the last decades has made possible the development of methods to quickly identify responsible pathogen using their DNA and without prior isolation. Long-read sequencing tehcniques like ONT make these methods even easier and more practical to identify strains quicker and with fewer reads.
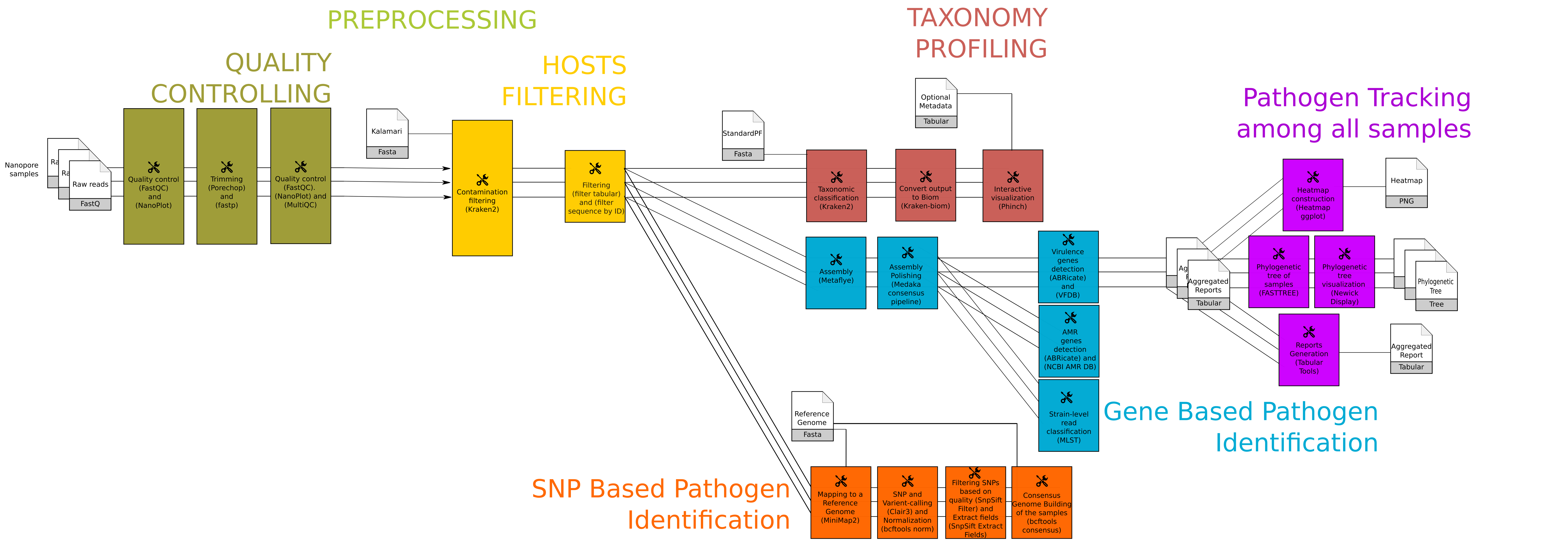
In this tutorial, using direct Nanopore sequencing data, we show how to
- agnostically detect pathogens (what exactly is this pathogen and what is the degree of its severity (its virulence factors) from data extracted directly (without prior cultivation) from a potentially contaminated sample (e.g. food like chicken, cow, etc.,) and sequenced using Nanopore
- compare different samples to track the possible source of contamination
This tutorial is offered in 2 different version:
- A short version, running already build workflows
- A long version, going step-by-step